Note
Click here to download the full example code
Use Fitroom with on-the-fly calculated spectra¶
Compare experimental spectrum to a combination of precomputed spectra
powered by the RADIS SpecDatabase class
Description¶
Functions to build a 2D, multislabs fitting room with a pure Python (Matplotlib) interface that displays:
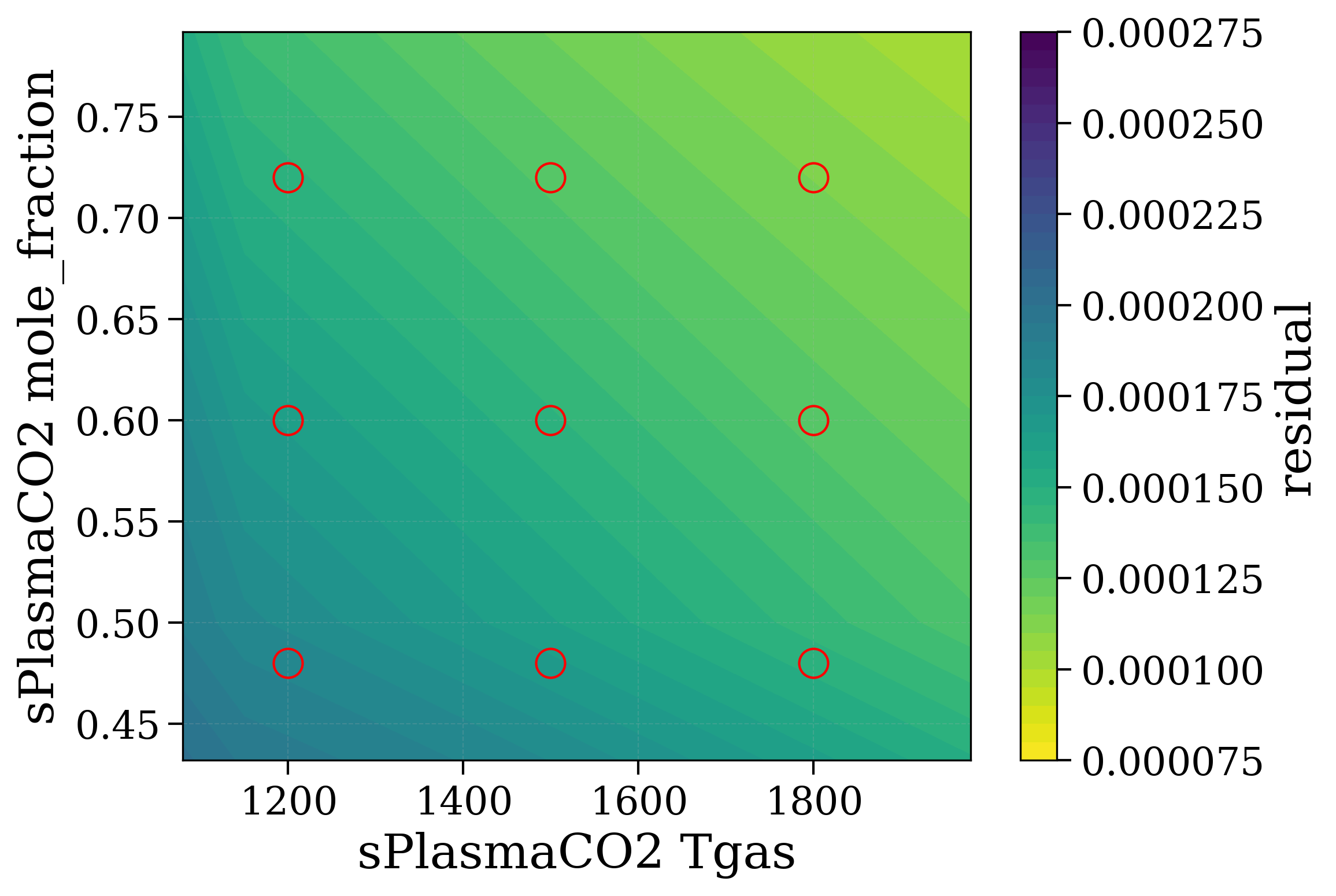
CaseSelectora window to select conditions along two axis (to calculate, or retrievefrom a database)
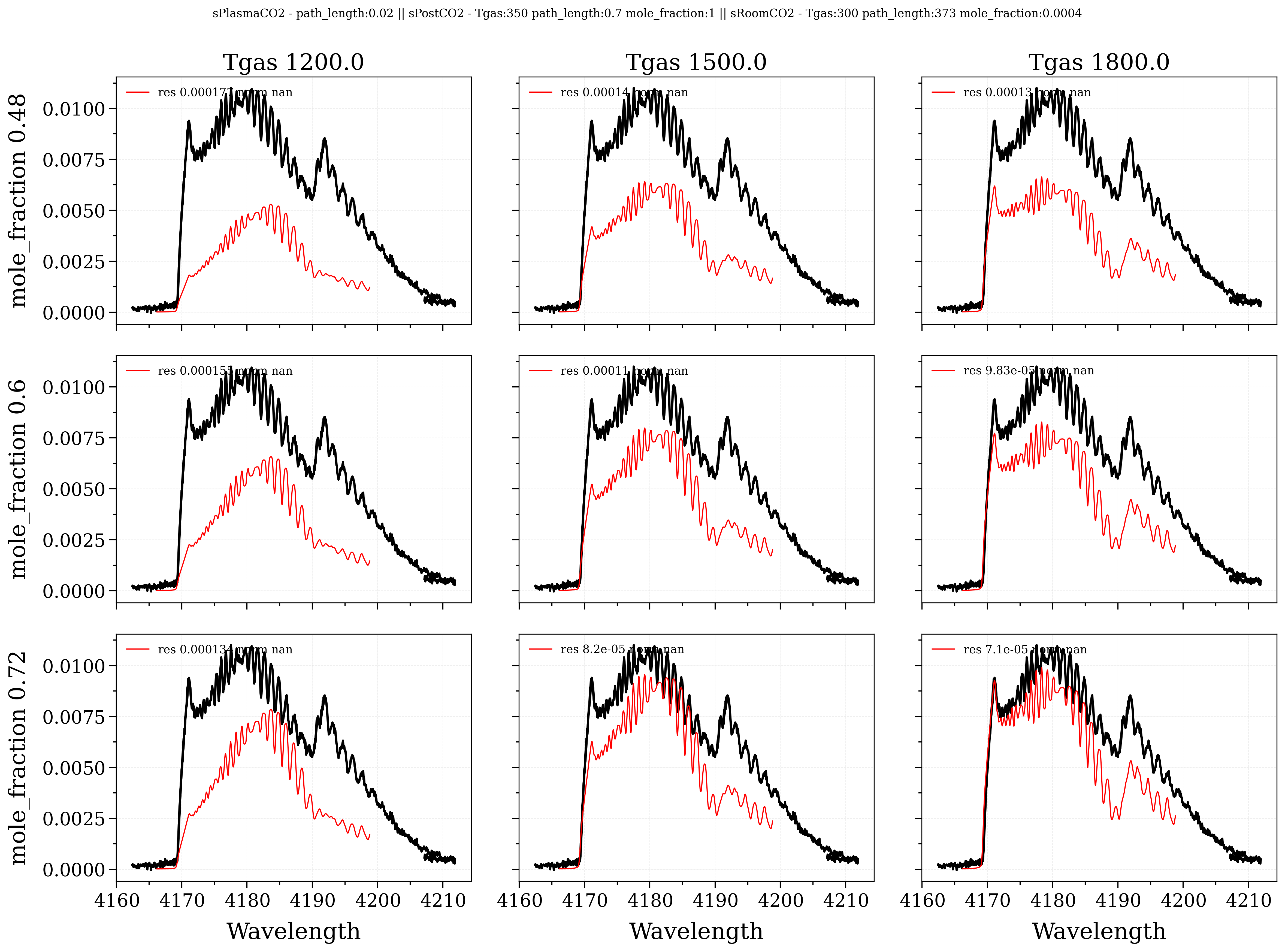
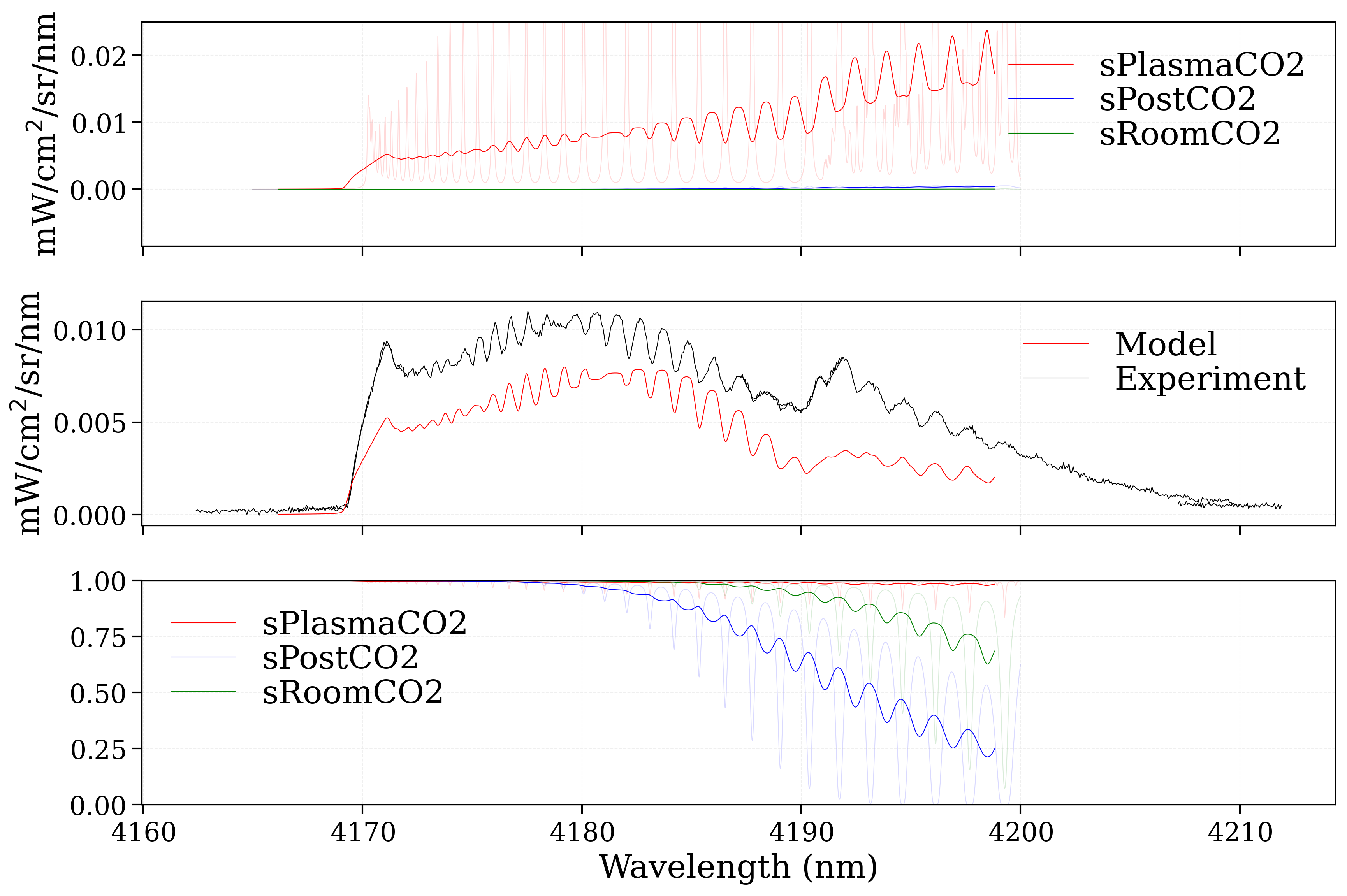
Grid3x3: a window to plot 9 spectra corresponding to left, right and center conditionsMultiSlabPlot: a window to decompose the center slab along the different slabs
This example calculates spectra on-fly.
Calculated spectra are stored in a “HITRAN_CO2_test_spec_database” folder
to be re-used, using the init_database() method
of SpectrumFactory
import warnings
from radis.los.slabs import SerialSlabs#, MergeSlabs
from radis.test.utils import setup_test_line_databases
import numpy as np
from numpy import linspace
from fitroom import CaseSelector
from fitroom import Grid3x3
from fitroom import MultiSlabPlot
from fitroom import SlabsConfigSolver
from fitroom import FitRoom
from fitroom import SlitTool
#from fitroom.tools import Normalizer
from radis import SpectrumFactory
from radis import Spectrum
from os.path import join, dirname
import matplotlib.pyplot as plt
plt.ion() # interactive mode; do not block figures
TEST_FOLDER_PATH = dirname(".")
def getTestFile(file):
''' Return the full path of a test file. Used by test functions not to
worry about the project architecture'''
return join(TEST_FOLDER_PATH, file)
wav_min = 4165
wav_max = 4200
setup_test_line_databases() # registers "HITRAN-CO2-TEST" for this example
sf2 = SpectrumFactory(wavelength_min=wav_min,
wavelength_max=wav_max,
cutoff=1e-25,
isotope=[1, 2],
medium='air')
sf2.load_databank('HITRAN-CO2-TEST')
sf2.init_database('HITRAN_CO2_test_spec_database')
Out:
Added HITRAN-CO2-TEST database in /home/docs/radis.json
Added HITRAN-CO-TEST database in /home/docs/radis.json
Added HITEMP-CO2-TEST database in /home/docs/radis.json
Added HITEMP-CO2-HAMIL-TEST database in /home/docs/radis.json
Using database: HITRAN-CO2-TEST
'HITRAN-CO2-TEST':
{'info': 'HITRAN 2016 database, CO2, 1 main isotope (CO2-626), bandhead: 2380-2398 cm-1 (4165-4200 nm)', 'path': ['/home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/radis/test/files/hitran_co2_626_bandhead_4165_4200nm.par'], 'format': 'hitran', 'parfuncfmt': 'hapi', 'levelsfmt': 'radis'}
Generating cache file /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/radis/test/files/hitran_co2_626_bandhead_4165_4200nm.h5 with metadata :
{'last_modification': 'Sat Oct 2 15:30:33 2021', 'wavenum_min': 2380.019436, 'wavenum_max': 2399.965532}
Reference databank (2380.31-2399.97cm-1) has 0 lines in range (2380.30-2400.31cm-1) for isotope 2. Change your range or isotope options
/home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/radis/misc/log.py:54: UserWarning: Reference databank (2380.31-2399.97cm-1) has 0 lines in range (2380.30-2400.31cm-1) for isotope 2. Change your range or isotope options
warn(msg)
HAPI version: 1.2.2.0
To get the most up-to-date version please check http://hitran.org/hapi
ATTENTION: Python versions of partition sums from TIPS-2021 are now available in HAPI code
MIT license: Copyright 2021 HITRAN team, see more at http://hitran.org.
If you use HAPI in your research or software development,
please cite it using the following reference:
R.V. Kochanov, I.E. Gordon, L.S. Rothman, P. Wcislo, C. Hill, J.S. Wilzewski,
HITRAN Application Programming Interface (HAPI): A comprehensive approach
to working with spectroscopic data, J. Quant. Spectrosc. Radiat. Transfer 177, 15-30 (2016)
DOI: 10.1016/j.jqsrt.2016.03.005
ATTENTION: This is the core version of the HITRAN Application Programming Interface.
For more efficient implementation of the absorption coefficient routine,
as well as for new profiles, parameters and other functional,
please consider using HAPI2 extension library.
HAPI2 package is available at http://github.com/hitranonline/hapi2
Database HITRAN_CO2_test_spec_database initialised in
*** Loading the database with 1 processor (0 files)***
<radis.tools.database.SpecDatabase object at 0x7f662e76fb50>
slbPlasmaCO2 = {'factory': sf2,
'Tgas': 1500,
# 'Tvib':1100,
# 'Trot':1200,
'path_length': 0.02,
'mole_fraction': 0.6,
}
slbPostCO2 = {'factory': sf2,
'Tgas': 350,
'path_length': 0.7,
#'mole_fraction':DynVar('sPlasmaCO2', 'mole_fraction'),
# TODO: line above doesnt work with precompute residual. Fix it!
'mole_fraction': 1,
}
slbRoomCO2 = {'factory': sf2,
'Tgas': 300,
'path_length': 373, # cm
'mole_fraction': 400e-6,
}
Slablist = {'sPlasmaCO2': slbPlasmaCO2,
'sPostCO2': slbPostCO2,
'sRoomCO2': slbRoomCO2}
def config(**slabs):
''' args must correspond to slablist. Indexes and order is important '''
return SerialSlabs(slabs['sPlasmaCO2'], slabs['sPostCO2'], slabs['sRoomCO2'])
# Sensibility analysis
slbInteractx = 'sPlasmaCO2'
xparam = 'Tgas'
slbInteracty = 'sPlasmaCO2'
yparam = 'mole_fraction'
xstep = 0.2
ystep = 0.2
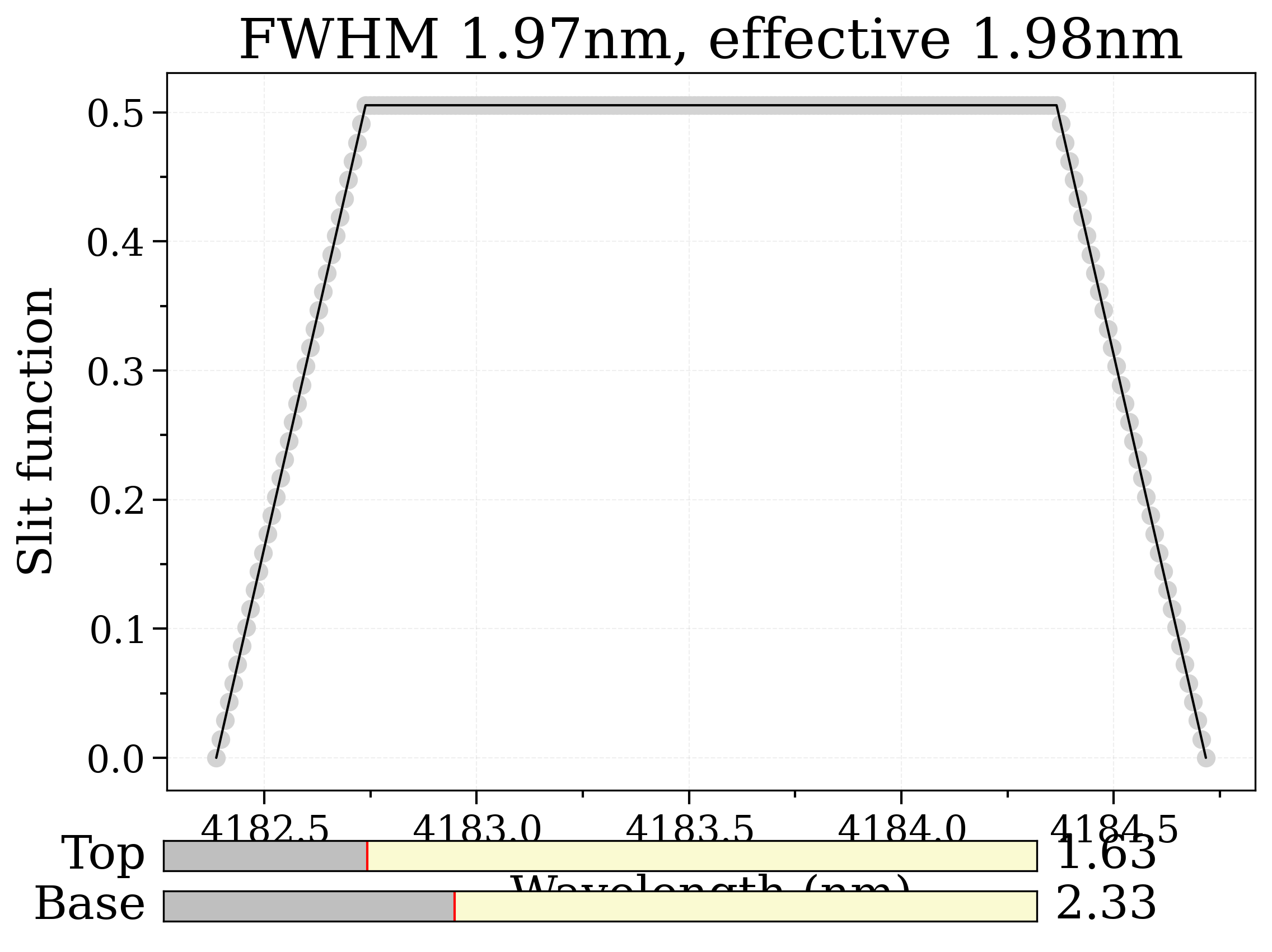
# Slit
slit = (1.63, 2.33)
# what to plot: 'radiance' or 'transmittance'
plotquantity = 'radiance'
unit = 'mW/cm2/sr/nm'
wunit = 'nm'
#unit = 'default'
s_exp = Spectrum.from_txt(getTestFile(r"measured_co2_bandhead_10kHz_30us.txt"),
quantity=plotquantity, wunit=wunit, unit=unit,
conditions={'medium': 'air'}).offset(-0.5, 'nm')
normalizer = None # Normalizer(4173, 4180, how='mean')
# -----------------------------------------------------------------------------
# NON USER PARAM PART
# -----------------------------------------------------------------------------
warnings.filterwarnings("ignore", "Using default event loop until function specific" +
"to this GUI is implemented")
fitroom = FitRoom(Slablist, slbInteractx, slbInteracty, xparam, yparam)
solver = SlabsConfigSolver(config=config, source='calculate',
s_exp=s_exp,
plotquantity=plotquantity, unit=unit,
slit=slit)
gridTool = Grid3x3(slbInteractx=slbInteractx, slbInteracty=slbInteracty,
xparam=xparam, yparam=yparam,
plotquantity=plotquantity, unit=unit,
normalizer=normalizer,
s_exp=s_exp)
slabsTool = MultiSlabPlot(plotquantity=plotquantity, unit=unit,
normalizer=normalizer,
s_exp=s_exp,
nfig=3)
selectTool = CaseSelector(xparam=xparam, yparam=yparam,
slbInteractx=slbInteractx, slbInteracty=slbInteracty,
nfig=1)
slitTool = SlitTool('nm') # getTestFile('slitfunction.txt'))
fitroom.add_tool(solver)
fitroom.add_tool(gridTool)
fitroom.add_tool(slabsTool)
fitroom.add_tool(selectTool)
# Map x, y
# -----------
xvar = Slablist[slbInteractx][xparam]
yvar = Slablist[slbInteracty][yparam]
xspace = linspace(xvar*(1-xstep), xvar*(1+xstep), 3)
yspace = linspace(yvar*(1+ystep), yvar*(1-ystep), 3)
gridTool.plot_3times3(xspace, yspace)
fitroom.add_tool(slitTool)
select_xspace = (xspace.min()*0.9, xspace.max()*1.1)
select_yspace = (yspace.min()*0.9, yspace.max()*1.1)
selectTool.ax.set_xlim(*select_xspace)
selectTool.ax.set_ylim(*select_yspace)
# Precompute residual for all points
# --------------
selectTool.precompute_residual(Slablist, # plotquantity='radiance',
xspace=np.linspace(300, 2000, 3),
yspace=np.linspace(0, 1, 3))
plt.show()
Out:
/home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/fitroom/selection_tool.py:131: UserWarning: Attempting to set identical left == right == 0 results in singular transformations; automatically expanding.
ax.set_xlim((xmin, xmax))
/home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/fitroom/selection_tool.py:132: UserWarning: Attempting to set identical bottom == top == 0 results in singular transformations; automatically expanding.
ax.set_ylim((ymin, ymax))
Adding SlabsConfigSolver
Adding Grid3x3
Adding MultiSlabPlot
Adding CaseSelector
/home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/radis/misc/warning.py:350: HighTemperatureWarning: HITRAN is valid for low temperatures (typically < 700 K). For higher temperatures you may need HITEMP or CDSD. See the 'databank=' parameter
warnings.warn(WarningType(message))
/home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/radis/tools/database.py:1224: UserWarning: Empty database
warn("Empty database")
No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1200.0 K
Trot 1200.0 K
Tvib 1200.0 K
isotope 1,2
mole_fraction 0.48
molecule CO2
overpopulation None
path_length 0.02 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used None
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding -1
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
/home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/json_tricks/encoders.py:367: UserWarning: json-tricks: numpy scalar serialization is experimental and may work differently in future versions
warnings.warn('json-tricks: numpy scalar serialization is experimental and may work differently in future versions')
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib1200K_Trot1200K.spec (0.1Mb)
loaded 20211002_Tvib1200K_Trot1200K.spec
0.06s - Spectrum calculated
No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 350 K
Trot 350 K
Tvib 350 K
isotope 1,2
mole_fraction 1
molecule CO2
overpopulation None
path_length 0.7 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 2002
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib350K_Trot350K.spec (0.1Mb)
loaded 20211002_Tvib350K_Trot350K.spec
0.05s - Spectrum calculated
No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 300 K
Trot 300 K
Tvib 300 K
isotope 1,2
mole_fraction 0.0004
molecule CO2
overpopulation None
path_length 373 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 2002
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib300K_Trot300K.spec (0.1Mb)
loaded 20211002_Tvib300K_Trot300K.spec
0.05s - Spectrum calculated
/home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/radis/misc/warning.py:350: HighTemperatureWarning: HITRAN is valid for low temperatures (typically < 700 K). For higher temperatures you may need HITEMP or CDSD. See the 'databank=' parameter
warnings.warn(WarningType(message))
No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1200.0 K
Trot 1200.0 K
Tvib 1200.0 K
isotope 1,2
mole_fraction 0.6
molecule CO2
overpopulation None
path_length 0.02 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 2002
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
Warning. File already exists. Filename is incremented
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib1200K_Trot1200K_1.spec (0.1Mb)
loaded 20211002_Tvib1200K_Trot1200K_1.spec
0.06s - Spectrum calculated
Spectrum found in database!
Spectrum found in database!
/home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/radis/misc/warning.py:350: HighTemperatureWarning: HITRAN is valid for low temperatures (typically < 700 K). For higher temperatures you may need HITEMP or CDSD. See the 'databank=' parameter
warnings.warn(WarningType(message))
No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1200.0 K
Trot 1200.0 K
Tvib 1200.0 K
isotope 1,2
mole_fraction 0.72
molecule CO2
overpopulation None
path_length 0.02 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 2002
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
Warning. File already exists. Filename is incremented
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib1200K_Trot1200K_2.spec (0.1Mb)
loaded 20211002_Tvib1200K_Trot1200K_2.spec
0.05s - Spectrum calculated
Spectrum found in database!
Spectrum found in database!
/home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/radis/misc/warning.py:350: HighTemperatureWarning: HITRAN is valid for low temperatures (typically < 700 K). For higher temperatures you may need HITEMP or CDSD. See the 'databank=' parameter
warnings.warn(WarningType(message))
No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1500.0 K
Trot 1500.0 K
Tvib 1500.0 K
isotope 1,2
mole_fraction 0.48
molecule CO2
overpopulation None
path_length 0.02 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 2002
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib1500K_Trot1500K.spec (0.1Mb)
loaded 20211002_Tvib1500K_Trot1500K.spec
0.05s - Spectrum calculated
Spectrum found in database!
Spectrum found in database!
/home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/radis/misc/warning.py:350: HighTemperatureWarning: HITRAN is valid for low temperatures (typically < 700 K). For higher temperatures you may need HITEMP or CDSD. See the 'databank=' parameter
warnings.warn(WarningType(message))
No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1500.0 K
Trot 1500.0 K
Tvib 1500.0 K
isotope 1,2
mole_fraction 0.6
molecule CO2
overpopulation None
path_length 0.02 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 2002
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
Warning. File already exists. Filename is incremented
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib1500K_Trot1500K_1.spec (0.1Mb)
loaded 20211002_Tvib1500K_Trot1500K_1.spec
0.05s - Spectrum calculated
Spectrum found in database!
Spectrum found in database!
/home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/radis/misc/warning.py:350: HighTemperatureWarning: HITRAN is valid for low temperatures (typically < 700 K). For higher temperatures you may need HITEMP or CDSD. See the 'databank=' parameter
warnings.warn(WarningType(message))
No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1500.0 K
Trot 1500.0 K
Tvib 1500.0 K
isotope 1,2
mole_fraction 0.72
molecule CO2
overpopulation None
path_length 0.02 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 2002
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
Warning. File already exists. Filename is incremented
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib1500K_Trot1500K_2.spec (0.1Mb)
loaded 20211002_Tvib1500K_Trot1500K_2.spec
0.06s - Spectrum calculated
Spectrum found in database!
Spectrum found in database!
/home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/radis/misc/warning.py:350: HighTemperatureWarning: HITRAN is valid for low temperatures (typically < 700 K). For higher temperatures you may need HITEMP or CDSD. See the 'databank=' parameter
warnings.warn(WarningType(message))
No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1800.0 K
Trot 1800.0 K
Tvib 1800.0 K
isotope 1,2
mole_fraction 0.48
molecule CO2
overpopulation None
path_length 0.02 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 2002
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib1800K_Trot1800K.spec (0.1Mb)
loaded 20211002_Tvib1800K_Trot1800K.spec
0.05s - Spectrum calculated
Spectrum found in database!
Spectrum found in database!
/home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/radis/misc/warning.py:350: HighTemperatureWarning: HITRAN is valid for low temperatures (typically < 700 K). For higher temperatures you may need HITEMP or CDSD. See the 'databank=' parameter
warnings.warn(WarningType(message))
No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1800.0 K
Trot 1800.0 K
Tvib 1800.0 K
isotope 1,2
mole_fraction 0.6
molecule CO2
overpopulation None
path_length 0.02 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 2002
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
Warning. File already exists. Filename is incremented
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib1800K_Trot1800K_1.spec (0.1Mb)
loaded 20211002_Tvib1800K_Trot1800K_1.spec
0.05s - Spectrum calculated
Spectrum found in database!
Spectrum found in database!
/home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/radis/misc/warning.py:350: HighTemperatureWarning: HITRAN is valid for low temperatures (typically < 700 K). For higher temperatures you may need HITEMP or CDSD. See the 'databank=' parameter
warnings.warn(WarningType(message))
No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1800.0 K
Trot 1800.0 K
Tvib 1800.0 K
isotope 1,2
mole_fraction 0.72
molecule CO2
overpopulation None
path_length 0.02 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 2002
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
Warning. File already exists. Filename is incremented
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib1800K_Trot1800K_2.spec (0.1Mb)
loaded 20211002_Tvib1800K_Trot1800K_2.spec
0.05s - Spectrum calculated
Spectrum found in database!
Spectrum found in database!
Adding SlitTool
/home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/fitroom/selection_tool.py:423: FutureWarning: elementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison
if xspace == 'database':
/home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/fitroom/selection_tool.py:425: FutureWarning: elementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison
if yspace == 'database':
(0s) 0.0% No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 300.0 K
Trot 300.0 K
Tvib 300.0 K
isotope 1,2
mole_fraction 0.0
molecule CO2
overpopulation None
path_length 0.02 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 2002
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
Warning. File already exists. Filename is incremented
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib300K_Trot300K_1.spec (0.0Mb)
loaded 20211002_Tvib300K_Trot300K_1.spec
0.04s - Spectrum calculated
Spectrum found in database!
Spectrum found in database!
No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 300.0 K
Trot 300.0 K
Tvib 300.0 K
isotope 1,2
mole_fraction 0.5
molecule CO2
overpopulation None
path_length 0.02 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 2002
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
Warning. File already exists. Filename is incremented
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib300K_Trot300K_2.spec (0.1Mb)
loaded 20211002_Tvib300K_Trot300K_2.spec
0.05s - Spectrum calculated
Spectrum found in database!
Spectrum found in database!
No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 300.0 K
Trot 300.0 K
Tvib 300.0 K
isotope 1,2
mole_fraction 1.0
molecule CO2
overpopulation None
path_length 0.02 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 2002
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
Warning. File already exists. Filename is incremented
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib300K_Trot300K_3.spec (0.1Mb)
loaded 20211002_Tvib300K_Trot300K_3.spec
0.05s - Spectrum calculated
Spectrum found in database!
Spectrum found in database!
(1s) 33.3% No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1150.0 K
Trot 1150.0 K
Tvib 1150.0 K
isotope 1,2
mole_fraction 0.0
molecule CO2
overpopulation None
path_length 0.02 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 2002
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib1150K_Trot1150K.spec (0.0Mb)
loaded 20211002_Tvib1150K_Trot1150K.spec
0.05s - Spectrum calculated
Spectrum found in database!
Spectrum found in database!
No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1150.0 K
Trot 1150.0 K
Tvib 1150.0 K
isotope 1,2
mole_fraction 0.5
molecule CO2
overpopulation None
path_length 0.02 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 2002
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
Warning. File already exists. Filename is incremented
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib1150K_Trot1150K_1.spec (0.1Mb)
loaded 20211002_Tvib1150K_Trot1150K_1.spec
0.06s - Spectrum calculated
Spectrum found in database!
Spectrum found in database!
No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1150.0 K
Trot 1150.0 K
Tvib 1150.0 K
isotope 1,2
mole_fraction 1.0
molecule CO2
overpopulation None
path_length 0.02 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 2002
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
Warning. File already exists. Filename is incremented
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib1150K_Trot1150K_2.spec (0.1Mb)
loaded 20211002_Tvib1150K_Trot1150K_2.spec
0.06s - Spectrum calculated
Spectrum found in database!
Spectrum found in database!
(1s) 66.7% No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 2000.0 K
Trot 2000.0 K
Tvib 2000.0 K
isotope 1,2
mole_fraction 0.0
molecule CO2
overpopulation None
path_length 0.02 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 2002
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib2000K_Trot2000K.spec (0.0Mb)
loaded 20211002_Tvib2000K_Trot2000K.spec
0.05s - Spectrum calculated
Spectrum found in database!
Spectrum found in database!
No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 2000.0 K
Trot 2000.0 K
Tvib 2000.0 K
isotope 1,2
mole_fraction 0.5
molecule CO2
overpopulation None
path_length 0.02 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 2002
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
Warning. File already exists. Filename is incremented
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib2000K_Trot2000K_1.spec (0.1Mb)
loaded 20211002_Tvib2000K_Trot2000K_1.spec
0.06s - Spectrum calculated
Spectrum found in database!
Spectrum found in database!
No spectrum found in database that matched given conditions.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 2000.0 K
Trot 2000.0 K
Tvib 2000.0 K
isotope 1,2
mole_fraction 1.0
molecule CO2
overpopulation None
path_length 0.02 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2400.3058 cm-1
wavenum_min 2380.3033 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-25 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/rad...
dlm_log_pG 0.1375350788016573
dlm_log_pL 0.20180288881201608
export_rovib_fraction False
folding_thresh 1e-06
hdf5_engine pytables
include_neighbouring_lines True
levelsfmt radis
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parfuncpath None
parsum_mode full summation
pseudo_continuum_threshold 0
truncation 50 cm-1
wavenum_max_calc 2400.3058 cm-1
wavenum_min_calc 2380.3033 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 2002
Information
----------------------------------------
chunksize None
db_use_cached True
export_lines False
export_populations None
load_energies False
lvl_use_cached True
total_lines 327
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
----------------------------------------
Warning. File already exists. Filename is incremented
Spectrum stored in HITRAN_CO2_test_spec_database/20211002_Tvib2000K_Trot2000K_2.spec (0.1Mb)
loaded 20211002_Tvib2000K_Trot2000K_2.spec
0.06s - Spectrum calculated
Spectrum found in database!
Spectrum found in database!
(2s) 100.0%
/home/docs/checkouts/readthedocs.org/user_builds/fitroom/envs/latest/lib/python3.8/site-packages/fitroom/selection_tool.py:500: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
plt.tight_layout()
Total running time of the script: ( 0 minutes 11.547 seconds)